AFWG shall not bear any responsibility for any content on such sites. Any link to a third-party site does not constitute an endorsement of the third party, their site or services. AFWG also makes no warranties as to the content of such sites.
Would you like to continue?
The pathogenic yeast cell with a polysaccharide capsule, Cryptococcus neoformans/C. gattii complex, causes a life-threatening infection called cryptococcosis. The natural habitats of Cryptococcus are soil with bird (eg, pigeon) droppings, decaying wood and the bark of certain trees. Recently, evidence showed that C. gattii could be isolated from some tropical trees, such as almond trees (Terminalia catappa), pottery trees (Ficus microcarpa) and cassia (Cassia grandis).
Cryptococcosis usually affects immunocompromised hosts, especially HIV patients, via the respiratory tract. After inhalation of either sexual or asexual spores, clinical symptoms develop and manifest in the lungs, blood and, finally, the brain and meninges, sometimes spreading to the lymph nodes and skin. Severe symptoms are seen in both humans and animals. Recently, laboratory surveillance studies have illustrated the variety of Cryptococcus, from the serotype to the molecular level (Table 1).
Genus and species | Serotype | Molecular type |
C. neoformans complex C. neoformans var. grubii
C. neoformans var. neoformans | A D | AFLP1/VNI AFLP1A, AFLP1B/VNII AFLP2/VNIV |
C. gattii complex C. gattii sensu stricto C. bacillisporus C. deuterogattii C. tetragattii C. decagattii | B C | AFLP4/VGI AFLP5/VGIII AFLP6/VGII AFLP7/VGIV AFLP10/VGIV |
| Hybrids | AD, AB, BD | AFLP3/VNIII |
Worldwide, about 1 million cases of cryptococcal meningitis occur among people with HIV/AIDS each year, resulting in nearly 625,000 deaths. This infection can be found globally, affecting sub-Saharan Africa, USA, Europe and Asia, including Thailand, where the burden of cryptococcal meningitis was estimated at 4.2/100,000 per year.
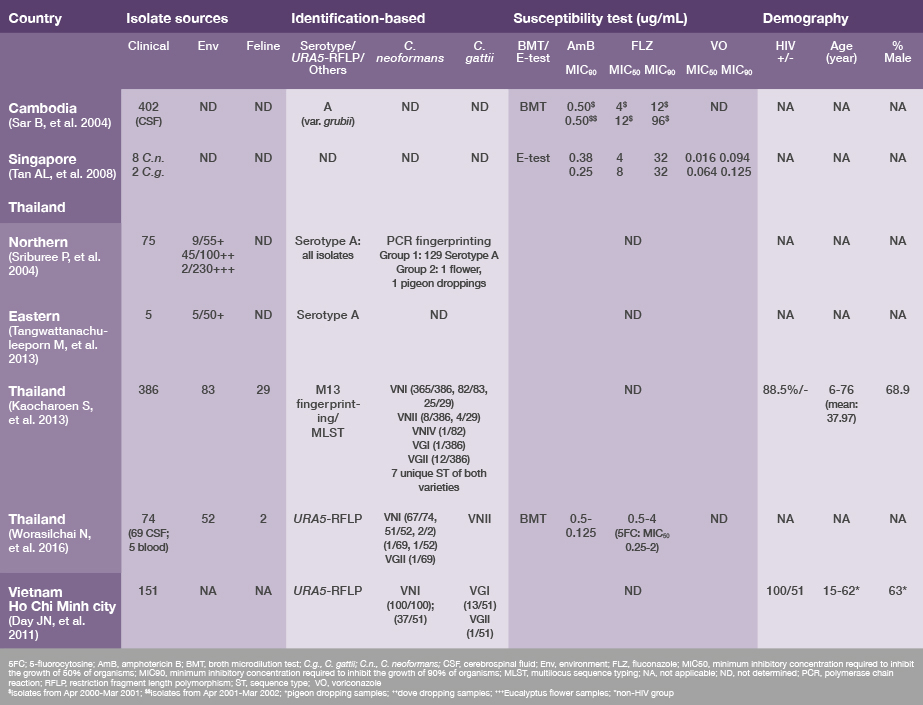
The majority of these cases in Thailand were in HIV-infected hosts and most of the isolates were C. neoformans var. grubii, serotype A/VNI, which is similar to findings in other countries. A summary of surveillance data from Southeast Asia is shown in Table 2, and includes cultures isolated from 1993 to 2011. Also similar to that seen in other countries, C. gattii complex has been on an upward trend in Southeast Asia from both clinical and environmental sources. A molecular study in Thailand showed a strong link between environmental and clinical strains for the VNI isolates, and similarities among the situations in Thailand and Vancouver (Canada), Australia and South America.
Moreover, a number of unique sequence types have been found in Thailand, which warrants further study in this region. Regarding the susceptibility profile, no resistance strains were presented in these studies, despite reports of resistance from other countries, such as Cambodia. Resistance should be monitored carefully, particularly since non–HIV-infected hosts have been reported.
For brain and meningeal infections, cerebrospinal fluid (CSF) and serum are the preferred specimens for laboratory diagnosis. India ink or nigrosin preparation is used to stain the background for simple direct examination. To increase the yield, one drop of sediment after 1,500 g/3,500 rpm for 10 minutes of centrifugation is recommended for CSF. Sputum, lymph node and tissue biopsy samples can also be used. The supernatant can be used for antigen detection either by lateral flow strip or latex agglutination test. To shorten the identification process, the specimens may be inoculated in the medium with phenoloxidase substrate (caffeic acid/mustard seed/cabbage seed) to induce the melanin production, which is a simple test to confirm the encapsulated yeasts as C. neoformans. Urease production of the encapsulated yeast is another rapid screening test for genus Cryptococcus. Concanavalin medium is one of the recommended tests that can be used to identify the variety. URA5-RFLP, multilocus sequence typing and other molecular techniques are also able to identify the genetic variation for both diagnostic and epidemiologic purposes.